Dimensionality Reduction (요약)
1. Feature extraction / Dimensionality reduction
- Given data points in $d$ dimensions
- Convert them to data points in $k(<d)$ dimensions
- With minimal loss of information
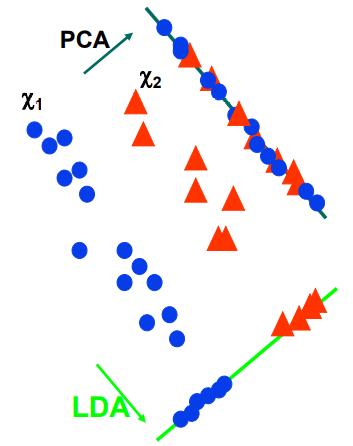
2. Principal Component Analysis (PCA)
- Find k-dim projection that best preserves variance
- Process
- compute mean vector $\mu$ and covariance matrix $\Sigma$ of original data $$\Sigma = \frac{1}{N}\sum_{i=1}^N (x_i - \mu)(x_i-\mu)^T $$
- Compute eigenvectors and eigenvalues of $\Sigma$ $$\Sigma v = \lambda v $$
- Select largest $k$ eigenvalues (also eigenvectors)
- Project points onto subspace spanned by them $$y = A(x - \mu)$$
- Eigenvector with largest eigenvalue captures the most variation among data $X$
- We can compress the data by using the top few eigenvectors (principal components)
- Feature vector $y_k$'s are uncorrelated (orthogonal)
3. Linear Discriminant Analysis (LDA)
- PCA vs. LDA
- PCA does not consider "class" information
- LDA consider "class" information
- PCA maximizes projected total scatter
- LDA maximizes ratio of projected between-class to projected within-class scatter
- Within-class scatter (want to minimize) $$\Sigma_w = \sum_{j=1}^c \frac{1}{N_c}\sum_{i=1}^{N_c} (x_i -\mu_c)(x_i -\mu_c)^T $$
- Between-class scatter (want to maximize) $$\Sigma_b = \frac{1}{c}\sum_{i=1}^c (\mu_i -\mu)(\mu_i -\mu)^T $$
- Compute eigenvectors and eigenvalues $$\frac{\Sigma_b}{\Sigma_c}v = \lambda v $$
댓글
댓글 쓰기